News
New Version of PhenDB
We are proud to present our updated version of PhenDB. While the frontend did not change that much, we added more functionality and up to date resources:
Faster, more robust trait prediction
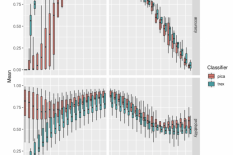
PICA has been updated to support python3 and use a more recent version of libSVM (via scikit-learn). To achieve this, the entire code was re-implemented which comes with additional changes in behaviour. The models are more lightweight (14 MB instead of 1.4 GB), training and prediction require less cpu time and prediction results are more robust towards incompleteness of input bins (Figure 1). We therefore decided to use a new name for this tool: phenotrex.
Support of EggNOG5
The EggNOG5 paper was published end of 2018, and now is our new standard database for annotation of bacterial bins. Although this represents an increase in Hidden-Markov-Models, we offer roughly the same speed for annotation due to improvements in our backend infrastructure.
Insights into predictions - SHAP
Since our re-implementation of PICA relies on scikit-learn, we have a lot of new possibilities to add functionality which makes use of the scikit-learn interface. One very useful new feature is the SHapely Additive exPlanations (SHAP) which provides insights into the individual importance of orthologous groups present or absent per sample to the final verdict of phenotrex.
Figure 1: Accuracy (5-fold CV) and probability of 10 models normalized to the range [0,1] for the two implementations of PICA: “pica” and (pheno)”trex” with simulated completeness and contamination in 5% steps.