Methods
About PhenDB
In recent years, metagenomics projects were often focused on taxonomic composition and diversity, which were used to explain the roles of uncultivated microbes in their community. However, due to lateral gene transfer (LGT) mechanisms and rapid mutation rates observed in many bacteria, assessing the presence or absence of a phenotypic trait in an operational taxonomic unit (OTU) by proxy of taxonomic position alone can be deceptive. Here, we provide a framework which gives a rough overview of phenotypic traits potentially present in a user-provided set of metagenomics bins on the basis of genomic content.
PhenDB is a publicly available resource able to screen previously assembled and binned metagenomes or genomes of cultivated isolates for 47 different bacterial traits. We use support vector machines (SVM) trained with manually curated datasets based on gene presence/absence patterns for trait prediction.
The PhenDB workflow
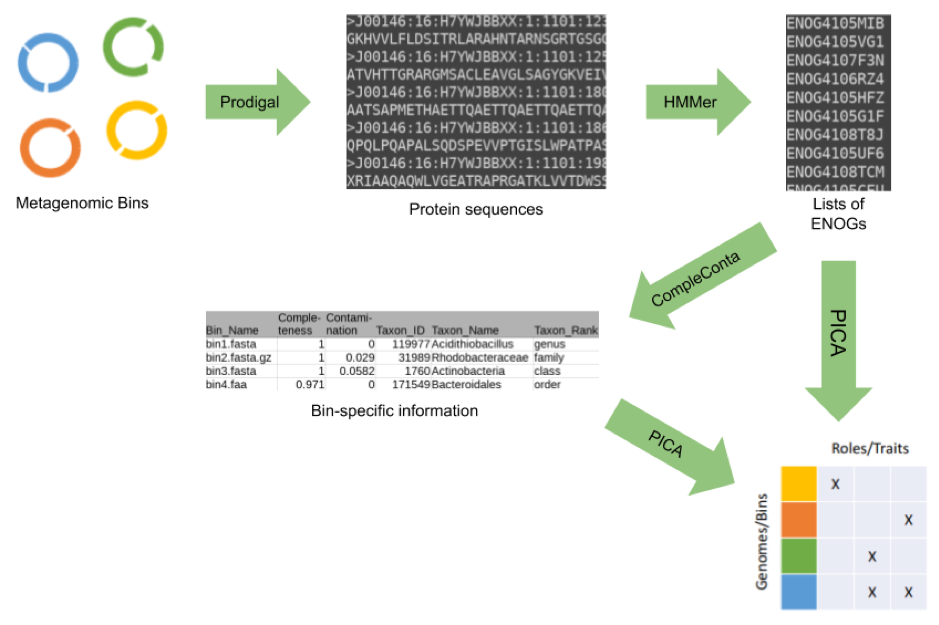
The PhenDB pipeline utilizes several applications to provide the user with trait predictions based on uploaded metagenomic bins.
Trait prediction is performed by utilizing a machine learning-based method called PICA as published in Feldbauer et al., 2015.
Together with the prediction, we also provide a rough estimation of bin completeness and contamination which PhenDB uses to gauge prediction quality. These values are derived from the presence and absence of 34 marker genes, extracted from the EggNOG annotation similar to CheckM. The completeness and contamination values are used to adjust the maximum accuracy achievable by each model individually. Note, that we use a marker gene set based on different HMM-profiles than CheckM does, therefore the values for highly contaminated genomes may vary.
Marker genes are also taxonomically classified using the bactNOG database. We use a lowest common ancestor algorithm as applied by MEGAN with majority rule to provide a taxonomic classification for each input bin. Note that contaminated bins may be unclassified because of the stringent cutoffs we apply.